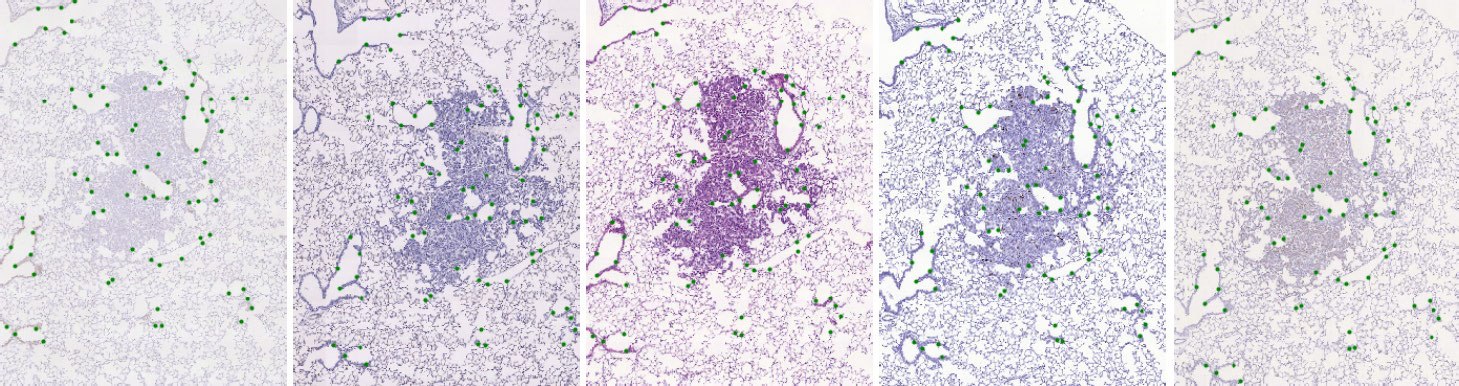
High-resolution (up to 40x magnification) whole-slide images of different types of tissue (lesions, lung-lobes, mammary-gland) were acquired - the original size of our images varies and goes from 15k x 15k up to about 50k x 50k pixels. The acquired images are organized in sets of consecutive tissue slices where each slice was stained by a different dye. The stains used are the following:
- clara cell 10 protein (Cc10)
- prosurfactant protein C (proSPC)
- hematoxylin and eosin (H&E)
- antigen KI-67 (Ki67)
- platelet endothelial cell adhesion molecule (PECAM-1, also known as CD31)
- human epidermal growth factor receptor 2 (c-erbB-2/HER-2-neu)
- estrogen receptor (ER)
- progesterone receptor (PR)
- cytokeratin
- podocin
We have in total 50+ histological sets. Each set is formed by whole slide images each corresponding to a cut of the same tissue stained with a different dye. For convenience, we provide downscaled versions of the images in 100%, 50%, 25%, 10% and 5% of the original size. The task is to register all images within sets among themselves. This forms a collection of meaningfully registration pairs.
Images
Short description of the particular tissue samples presented in this dataset.
Lesion tissue - Unstained adjacent 3μm formalin-fixed paraffin-embedded sections were cut from the blocks and stained with Hematoxylin and Eosin (H&E) or by immunohistochemistry with a specific antibody for CD31, proSPC, CC10 or Ki67. Images of three mice lung lesions (adenoma or adenocarcinoma) were acquired with a Zeiss Axio Imager M1 microscope (Carl Zeiss, Jena, Germany) equipped with a dry Plan Apochromat objective (numerical aperture NA=0.95, magnification 40×, pixel size 0.174 μm/pixel).
Lung lobes - The images of the four whole mice lung lobes correspond to the same set of histological samples as the lesion tissue. They were also acquired with a Zeiss Axio Imager M1 microscope (Carl Zeiss, Jena, Germany) equipped with a dry EC Plan-Neofluar objective (NA=0.30, magnification 10×, pixel size 1.274 μm/pixel).
Mammary glands - The sections are cuts from two mammary glands blocks stained with H&E (even sections) and alternatively, with an antibody against the estrogen receptor (ER), progesterone receptor (PR), or Her2-neu (odd sections). The images were acquired with the same microscope and set of acquisition parameters as the mice lung lobes. They were also acquired with a Zeiss Axio Imager M1 microscope (Carl Zeiss, Jena, Germany) equipped with a dry EC Plan-Neofluar objective (NA=0.30, magnification 10×, pixel size 2.294 μm/pixel).
COAD - The COlon ADenocarcinoma (COAD) set assembles series of histological sections from colon cancer samples, scanned with a 3DHistec Pannoramic MIDI II scanner at 10x magnification, for a resolution of 0.468 microns/pixel with a white-balance set to auto. Each series consists of one H&E histopathology section (first cut) followed by a variable number (4-7) of immunohistopathology sections with stains for the immune response (including CD4, CD68) and hypoxia. Due to technical reasons (e.g., some stains needed to be re-processed), the order of the sections is not guaranteed. Nevertheless, the sections are from a small volume of a tissue block.
Mice Kidney tissue - The set consists of resected healthy mice kidneys which show high similarity to human kidneys. We used nine consecutive whole slide images having similar tissue structures. Whole slides were digitized with a NanoZoomer 2.0HT scanner (Hamamatsu) and a 20× objective lens. The images were each roughly of 37k × 30k pixels size. Each image was dyed with one of the three stains - PAS, SMA or CD31, such that every alternate slide is a PAS image.
Gastric mucosa and gastric adenocarcinoma tissue - Surgical material from patients with a histologically verified diagnosis (gastric adenocarcinoma) were used for routine staining with Hematoxylin and Eosin (H&E) or for immunophenotyping. IHC-staining for LMP-1 protein (Dako, clone CS.1-4) were used for Epstein-Barr virus (EBV) identification. The study of the cellular composition of the tumour tissue infiltrate was performed by immunohistochemical staining on the markers CD4 (clone 4B12), CD8 (clone C8/144B), CD68 (clone PG-M1) and CD1a (clone O10). Deparaffinization and antigen recovery was performed by using Thermo Dewax and HIER Bufer L, pH6 buffer. The preparations were studied with a Leica DM LB2 microscope by two independent researchers.
Breast tissue - Unstained adjacent 3μm formalin-fixed paraffin-embedded sections were cut from the blocks and stained with Hematoxylin and Eosin (H&E) and with immunohistochemistry (IHC) with an antibody against the estrogen receptor (ER), progesterone receptor (PR), and Her2-neu.
Kidney tissue - Unstained adjacent 3μm formalin-fixed paraffin-embedded sections were cut from the glomerulopathies blocks and stained with Hematoxylin and Eosin (H&E) and PAS, Masson and Methenamine.
Summary
| Name | Tissue | Scanner | Magnitude | Resolution [µm/pixel] | Avg. size [pixels] |
| lung-lesion_ | Lung lesion | Zeiss Axio Imager M1 | 40x | 0.174 | 18k×15k |
| lung-lobes_ | Whole mice lung lobes | Zeiss Axio Imager M1 | 10x | 1.274 | 11k×6k |
| mammary-glands_ | Mammary glands | Zeiss Axio Imager M1 | 10x | 2.294 | 12k×4k |
| mice-kidney_ | Mice kidney | NanoZoomer 2.0HT (Hamamatsu) | 20x | 0.227 | 37k×30k |
| COAD_ | COlon ADenocarcinoma (colon cancer) | 3DHistec Pannoramic MIDI II | 10x | 0.468 | 60k×50k |
| gastric_ | Gastric mucosa and gastric adenocarcinoma tissue fragments | Leica Biosystems Aperio AT2 | 40x | 0.2528 | 60k×75k |
| breast_ | Human breast | Leica Biosystems Aperio AT2 | 40x | 0.2528 | 65kx60k |
| kidney_ | Human kidney | Leica Biosystems Aperio AT2 | 40x | 0.2528 | 18kx55k |
Landmarks
We have marked significant structures in the tissue with landmarks which are spread approximately uniformly over the tissue. Landmarks were manually identified in each image, with correspondences within each set, which allows us to validate the geometric registration accuracy between any two images in each set.
To evaluate training and testing performance, we do not provide all landmarks for each set (according to the cover file specifying the registrations pairs) and some will be kept private until the challenge is closed. Landmarks for training images will be freely available from the beginning of the challenge or are available already. The test landmarks will be used for evaluation only, on a server side. The evaluation framework is freely available.
The landmarks have standard ImageJ structure and coordinate frame (the origin [0, 0] is the top left pixel of the image plane).
The landmark file looks like this:
| ,X,Y 1,226,173 2,256,171 3,278,182 4,346,207 ... |
For handling landmark annotations or creating new annotations please follow these instructions, https://borda.github.io/dataset-histology-landmarks.
Directory Structure
The dataset is organised in sets and scales. The landmarks are in the CSV file with the same basename name as the image.
| DATASET |- lesions_1 | |- scale-5pc | | |- 29-041-Izd2-w35-CD31-3-les1.jpg | | |- 29-041-Izd2-w35-CD31-3-les1.csv | | |- 29-041-Izd2-w35-CD31-3-les1.jpg | | |- 29-041-Izd2-w35-CD31-3-les1.csv | | | ... | | |- 29-041-Izd2-w35-CD31-3-les1.jpg | | '- 29-041-Izd2-w35-CD31-3-les1.csv | |- scale-10pc | | ... | '- scale-100pc | |- 29-041-Izd2-w35-CD31-3-les1.png | |- 29-041-Izd2-w35-CD31-3-les1.csv | | ... | |- 29-041-Izd2-w35-CD31-3-les1.png | '- 29-041-Izd2-w35-CD31-3-les1.csv |- lesions_2 | ... '- mammary-gland_2 |
How to Download the Dataset
First, participants need to read and by downloading they accept the Licence terms. After that, the participants need to create an account on grand-challenge.org website (see Login/Register in the top right corner). After successful registration, they need to join the challenge before downloading the training dataset. The ‘Join’ link on the left may be used. After your request is completed, the dataset may be downloaded from the ‘Download’ link on the left.
Data Usage Agreement
You are free to use the data provided in the frame of this challenge in your own research work, provided that you acknowledge the source of the data and cite the references [1,2,3,4,5] whenever appropriate. This dataset is made available under the following licence: CC-BY-NC-SA.
Acknowledgement
The lesions, lung-lobes and mammary-gland images were provided by Prof. Carlos Ortiz de Solórzano and Dr. Arrate Munoz Barrutia, Center for Applied Medical Research (CIMA), University of Navarra, Pamplona Spain [1,2]. The mice kidney images were provided by Prof. Peter Boor and Dr. Barbara M. Klinkhammer, Institute of Pathology, University Hospital Aachen, RWTH Aachen University [3]. The colorectal cancer images were provided by Dr. Rudolf Nenutil (Masaryk Memorial Cancer Institute Brno), and Dr. Eva Budinska and Dr. Vlad Popovici (Masaryk University Brno) and were collected under grant nr.16-31966A by Ministry of Health of the Czech Republic. Gastric mucosa and gastric adenocarcinoma tissue images were provided by Prof. Pavel G. Malkov, Dr. Natalya V. Danilova, Dr. Nina A. Oleynikova and Ilya A. Mikhailov, Department of Pathology, Lomonosov Moscow State University [4]. The kidney and breast cancer whole slide images were provided by Dr. Gloria Bueno and Dr. Oscar Deniz from Grupo VISILAB, Universidad de Castilla-La Mancha (UCLM). The images were obtained and prepared thanks to the AIDPATH European project [5] coordinated by UCLM.
Bibliography
- Borovec J, Munoz-Barrutia A, Kybic J. Benchmarking of Image Registration Methods for Differently Stained Histological Slides. 2018 25th IEEE International Conference on Image Processing (ICIP). 2018. doi:10.1109/icip.2018.8451040
- Fernandez-Gonzalez R, Jones A, Garcia-Rodriguez E, Chen PY, Idica A, Lockett SJ, et al. System for combined three-dimensional morphological and molecular analysis of thick tissue specimens. Microsc Res Tech. 2002;59: 522–530.
- Gupta L, Klinkhammer BM, Boor P, Merhof D, Gadermayr M. Stain independent segmentation of whole slide images: A case study in renal histology. 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018). 2018. doi:10.1109/isbi.2018.8363824
- Mikhailov I, Danilova N, Malkov P. The immune microenvironment of various histological types of ebv-associated gastric cancer. Virchows Archiv. 2018;473: no. s1 doi:10.1007/s00428-018-2422-1
- Bueno G., Deniz O., AIDPATH: Academia and Industry Collaboration for Digital Pathology http://aidpath.eu/?page_id=279
- J. Borovec et al., "ANHIR: Automatic Non-rigid Histological Image Registration Challenge," in IEEE Transactions on Medical Imaging.