Welcome to the Automatic Non-rigid Histological Image Registration (ANHIR) challenge website. This challenge is a part of the IEEE International Symposium on Biomedical Imaging (ISBI) 2019.
This challenge aims at the automatic nonlinear image registration of 2D microscopy images of histopathology tissue samples stained with different dyes. The task is difficult due to non-linear deformations affecting the tissue samples, different appearance of each stain, repetitive texture, and the large size of the whole slide images.

Examples of stained tissues.
Background¶
In Digital Pathology, one of the most simple and yet most useful features is to visually compare successive tissue sections (slices) which require aligning all images to a common frame. Other related applications requiring image alignment are 3D reconstruction, image fusion, etc. Image alignment enables the pathologist to evaluate the histology and expression of multiple markers for a patient in a single area. In addition, due to tissue processing and pre-analytical steps, sections may suffer from non-linear deformations. That is, they will stretch and change shape from section to section. Currently, there are only a few automatically alignment tools that are able to handle large images with sufficient accuracy and reasonable processing time.
The proposed challenge focuses on comparing the accuracy and the speed of automatic non-linear registration methods on a set of large images from the same tissue samples but stained with different biomarkers. In our challenge, the registration accuracy will be evaluated using manually annotated landmarks. We will also estimate the method robustness by computing how many times the performed registration improves the final image alignment. As an auxiliary criterion, the computation time will be also measured. We require that all methods run fully automatically, with no interaction and no image specific parameters (such as placing key-points or tuning parameters for some special images).
Participation¶
Teams, as well as individuals, are invited to participate and compare their image registration methods on this challenging dataset. We strongly suggest that the methods are made publicly available. For more information see Dataset and Evaluations pages. We provide a benchmark framework - https://borda.github.io/BIRL which contains several useful scripts and the benchmark implementation.
Data summary¶
High-resolution (up to 40x magnification) whole-slide images of tissues (lesions, lung-lobes, mammary-glands) were acquired - the original size of our images is up to 100k x 200k pixels. The acquired images are organized in sets of consecutive sections where each slice was stained with a different dye and any two images within a set can be meaningfully registered.
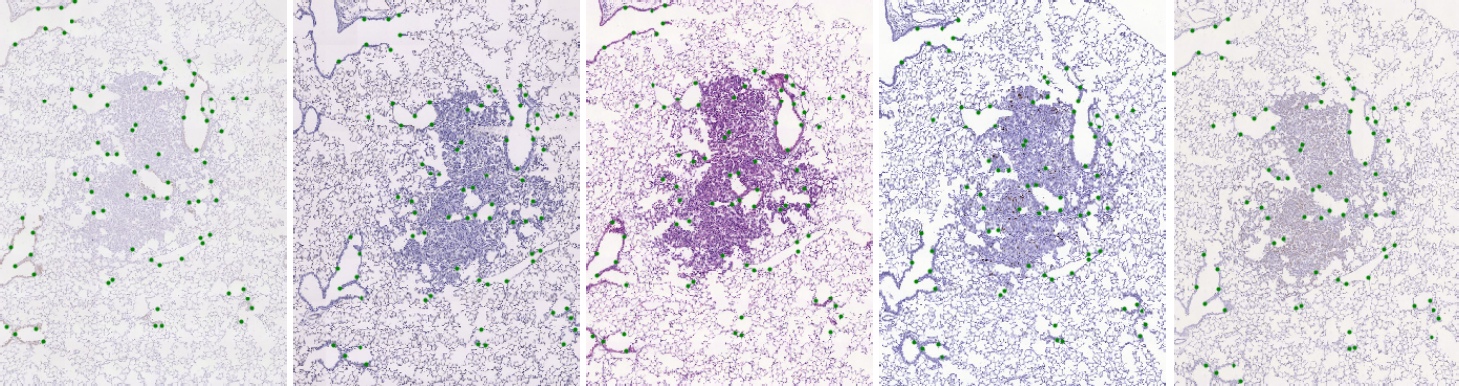
The task is to register the images based on the visible tissue. The background can be ignored. We have annotated significant structures in the tissue with landmarks which are approximately uniformly spread over the tissue. Each landmark position is known in all images from the same set. Landmarks are used to evaluate the quality of the registration.

An example of lung lesion tissue using several stains, with landmarks marked in green.
Timetable¶
- Datasets available for download [end of December 2018]
- Submission of results possible (see Evaluation page) [February 2019]
- Release a subset of training landmarks [February 2019]
- Release full-set of test landmarks [17th March 2019]
- Submission system closed [31st March 2019 at 23:59 (PDT)]
- Paper/report deadline [2nd April 2019 at 23:59 (PDT)]
- Invitation to present at the workshop sent [4th April 2019]
- Results announced at the conference [11th April 2019]
Sponsors¶




If you would like to sponsor this challenge, please send an email to kybic[at]fel.cvut.cz.
Presentation & Publications¶
A half-day workshop is planned to be held as part of the IEEE International Symposium on Biomedical Imaging (ISBI’19) where the final results of the challenge competition will be presented. Participating teams exceeding minimum performance requirements (with 85% robustness and rTRE lower than 1%) or ranked top 10 will be asked to submit a short paper (minimum 1-page and maximum 4-pages) in standard conference format and they will be invited to present their approach at the workshop (time scope will be adjusted according to the number of presenters).
A comprehensive report of this challenge and its results has been reported in an overview manuscript that we submitted to a high-quality peer-reviewed journal, such as IEEE Transactions in Medical Imaging, Medical Image Analysis, or Journal of Medical Imaging. Teams exceeding minimum performance requirements will be invited to participate as co-authors.
- Borovec, J., Kybic, J., et al. (2020). ANHIR: Automatic Non-rigid Histological Image Registration Challenge. IEEE Transactions on Medical Imaging (TMI). doi: 10.1109/TMI.2020.2986331
- Borovec, J. (2019). BIRL: Benchmark on Image Registration methods with Landmark validation. arXiv preprint arXiv:1912.13452.
- Borovec, J., Munoz-Barrutia, A., & Kybic, J. (2018). Benchmarking of image registration methods for differently stained histological slides. In IEEE International Conference on Image Processing (ICIP) (pp. 3368–3372), Athens. DOI: 10.1109/ICIP.2018.8451040